Scientists found 5' End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding
Mammalian cells contains NAD+-capped mRNAs and degraded by DXO family enzymes and scientists found that 5' end Nicotinamide Adenine Dinucleotide cap in human cells promotes RNA decay through DXO-Mediated deNADding.
5' End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay
Source
Dept. Cell Biology and Neuroscience, Rutgers University, Piscataway
Highlights
- Mammalian cells contains NAD+-capped mRNAs
- DXO family proteins are "deNADding" enzymes which remove the NAD+ cap from an mRNA
- A 5’ NAD+ cap promotes DXO-mediated RNA decay
- Intronic snoRNAs can be NAD+ capped and degraded by DXO in mammalian cell
- Highly efficient 5' capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase
The chemical nature of the 5′ end of RNA is a key determinant of RNA stability, processing, localization, translation efficiency. It provides a layer of “epitranscriptomic” gene regulation. The 5’ ends of eukaryotic mRNAs are cotranscriptionally modified by the addition of N7 methyl guanosine which fulfills multiple functions like the stability of mRNA, efficient translation and export of RNA. There are some derivative of m7G reported class of small U-rich noncoding RNAs that contain trimethylated, m2,2,7G cap m6A provides stability by Dcp2 decapping activity. Ribose of m7G 1st nucleotide and second nucleotide can be also methylated at 2nd position O2´-methylation of ribose also found in RNA.
Megerditch Kiledjian group showed other type of RNA capping and its role. When in place of m7G other molecules incorporated in the RNA of prokaryotes (PK) or eukaryotes (EK) is known as non-canonical initiating nucleotide (NCIN). It had long presumed that capped RNA present only in the EK RNA.
It has recently reported that capping is also present in bacteria. RNA products like some regulatory RNA gatY, gadY, RNAI in bacteria contain NAD cap structure. NAD+ can protect the RNA from 5’ end decay by the bacterial RppH and RNaseE.
NAD Capped RNA can be Degraded by NudC
NudC enzyme known to degrades NAD capped RNA. NAD+, NADH, and dpCoA are incorporated into RNA during transcription initiation by bacterial RNA polymerase and EK RNA pol II in vitro. Based on these reports authors studied that NAD+ caps can be added to RNA 5’ ends in mammalian cells.
They checked the presence of NAD capped RNA in mammalian cells. They used the NAD capture technique to isolate NAD capped RNA from mammalian cells. The role of the m7G cap in mRNA splicing and polyadenylation prompted them to analyze whether NAD+-capped RNAs are spliced and polyadenylated. They observed 20%–50% of the NAD+-capped mRNAs contain a polyadenylated tail and can be spliced. As previously reported that NAD cap in bacteria can protect RNA from exonuclease cleavage and NudC is an enzyme that degrades NAD capped RNA.
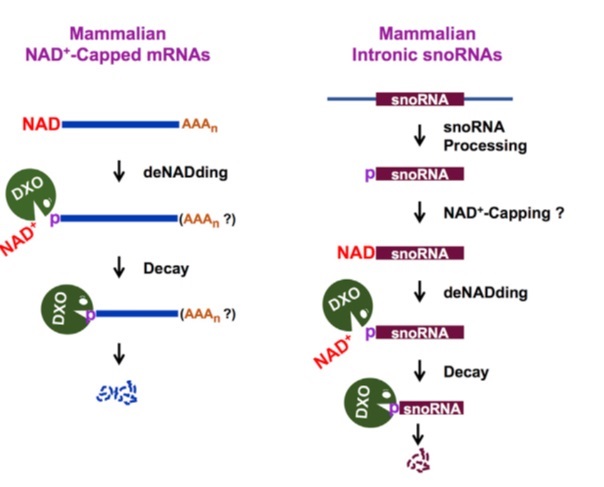
Next they checked that is there any enzyme in the mammalian cell which can degrades NAD cap. Mammalian cells contain many hydrolases like Dcp2, Nudt2, Nudt3 which hydrolyze mostly m7G capped RNA (completely capped RNA). DXO family proteins have pyrophosphohydrolase, Decapping, and 5′-3′ exoribonuclease activity which hydrolyze incompletely capped, unmethylated capped RNA. Analogous to removal of the entire cap structure, DXO might also remove an NAD+ modification on the 5’ end of RNAs. DXO might also remove an NAD+ modification on the 5’ end of RNAs
DXO hydrolyzed the m7G-capped RNA releasing the cap structure product. Surprisingly, DXO exhibited robust preferential activity on NAD+-capped RNA with an approximate 6-fold greater efficiency. This activity was compromised by the E234A or K255Q catalytic site mutant. They concluded DXO acts on NAD+-capped RNA releasing NAD+ (NppA) and pRNA and this process referred as ‘‘deNADding.’’
Also, m7G-capped RNA exhibited comparable stabilities in Con-KO cells, DXO-KO cells, and Dcp2-KO cells. DXO modulates the stability of NAD+-capped RNA, but not m7G-capped RNA in HEK293T cells. As well as luciferase experiments showed similar results. NAD+-capped luciferase mRNA is less stable than the same uncapped mRNA indicating the NAD+ cap promotes RNA decay. NAD+-capped luciferase mRNA was more stable in DXO-KO cells relative to control cells.
Translation capacity also decreased of NAD+ capped RNA. DXO preferentially targets a subset of snoRNAs and scaRNAs for deNADding. Fungal Rai1 and Dxo1 proteins are “deNADding” enzymes that suggests experiments.
Another report indicated that mtRNAPs serve as both sensors and actuators in coupling cellular metabolism to mitochondrial transcriptional outputs, sensing NAD+ and NADH levels and adjusting transcriptional outputs accordingly.

Journal Reference
Jiao X, Doamekpor SK, Bird JG, et al. 5' End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding. Cell. 2017;168(6):1015–1027.e10. doi:10.1016/j.cell.2017.02.019
