Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry
Antibody-Dependent Enhancement occurs when antibodies facilitate viral entry into host cells and enhance viral infection. ADE is found to be associated with a variety of viruses, mostly with flaviviruses. Detailed molecular mechanism for Antibody-Dependent Enhancement of coronavirus entry is still unknown.
Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry
Source
- University of Minnesota, USA
Highlights
- Coronavirus spike protein mediates viral entry into cells
- Neutralizing monoclonal antibody (MAb), which targets the receptor binding domain (RBD) of MERS coronavirus spike mediate virus entry to cells.
- ADE occurs only at intermediate MAb dosages.
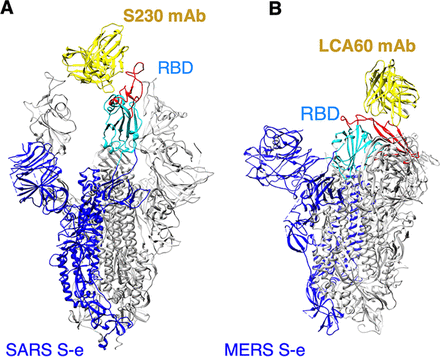
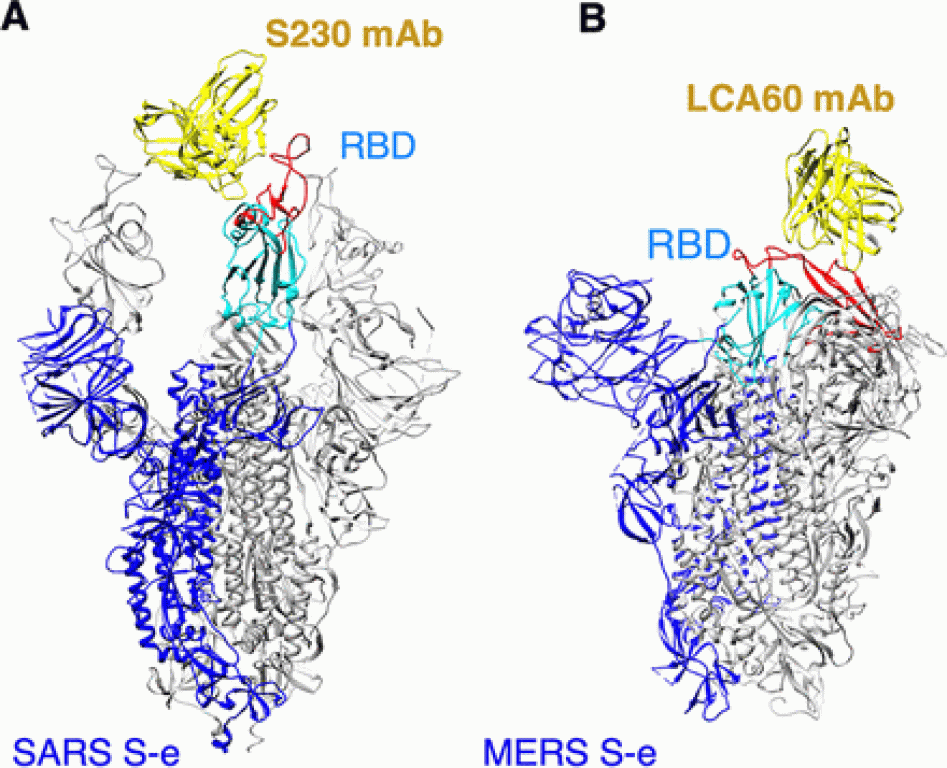
Figure: Structures of coronavirus spike proteins complexed with the antibody.
Corona Viruses
Coronaviruses are a family of large, positive-stranded, and enveloped RNA viruses. Two highly pathogenic coronaviruses, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome coronavirus (MERS-CoV), cause lethal infections in their spike protein guides entry into host cells. Spike is trimeric and contains three receptor binding subunit (S1) and a trimeric membrane fusion stalk (S2). SARS-CoV and MERS-CoV recognize angiotensin-converting enzyme 2 (ACE2) and dipeptidyl peptidase 4 (DPP4), respectively, as their viral receptors. The S1 subunit contains a receptor-binding domain (RBD) that mediates receptor recognition. Binding of RBD to the viral receptor can stabilize it in the standing-up state and triggers the spike to undergo further conformational changes, allowing host proteases to cleave first at the S1/S2 boundary and then within S2 (S2’ site). After two protease cleavages, S1 dissociates and S2 undergoes a dramatic structural change to fuse host and viral membranes and establish infection.
Antibody-Dependent Enhanced Entry
Antibody-Dependent Enhancement (ADE) occurs when antibodies facilitate viral entry into host cells and enhance viral infection. ADE is found to be associated with a variety of viruses, mostly with flaviviruses. In the case of dengue, HIV, and Ebola virus, antibodies produced after the first infection do not neutralize virus during secondary infection by another serotype of the virus, they instead bind to the virus and then to the IgG Fc receptors on immune cells and mediate viral entry into these cells. It has long been known that immunization of cats with feline coronavirus spike leads to worsened future infection due to ADE. However, detailed molecular mechanisms for ADE of coronavirus entry are still unknown. In this study, the authors revealed a novel mechanism for ADE in which fully neutralizing antibodies mimic the function of the viral receptor in mediating viral entry into Fc receptor-expressing cells.
Yushun Wan at College of Veterinary Medicine, University of Minnesota, USA carried out this study to investigate ADE of coronavirus entry. They used the mersmab1 antibody which was discovered in the same lab and has a strong binding affinity for MERS-CoV RBD and efficiently neutralizes MERS-CoV entry by outcompeting DPP4. They showed although RBD and MERS-CoV spike ectodomain (S-e) both bind to Mersmab1, S-e bound to Mersmab1 more tightly than the RBD, likely due to the trimeric state of S-e. In absence of Mersmab1, S-e bind to DPP4 and in its presence bind to CD32A (Fc receptor) cell surface receptor. MERS-CoV S-e binding to DPP4 is stronger than Mersmab1 mediated indirect binding of MERS-CoV S-e RBD to the Fc receptor.
They further investigated Mersmab1 mediates MERS-CoV entry into different Fc receptors (CD16A, CD32A, and CD64A)-expressing cells and macrophages (having a mixture of Fc Receptor) using pseudovirus entry assay. They found that in the absence of Mersmab1, MERS-CoV pseudoviruses could not enter Fc receptor-expressing cells but entered DPP4-expressing cells. On the other hand, in the presence of Mersmab1, MERS-CoV pseudoviruses entered CD32A-expressing cells and macrophages but were effectively blocked in DPP4 expressing cells. To expand these observations, they investigated the ADE of SARS-CoV entry. Using their previously identified SARS-CoV RBD-specific Mab (33G4) which binds to the ACE2-binding region of SARS-CoV RBD, they showed that 33G4 mediate SARS-CoV pseudovirus entry into CD32A expressing cells but blocked SARS-CoV pseudovirus entry into ACE2-expressing cells. Collectively these results suggest, both the MERS and SARS-CoV RBD-specific Mab can mediate entry into Fc receptor-expressing human cells while inhibiting entry into viral-receptor expressing human cells.
Molecular Mechanism
Next, to understand the molecular mechanism of ADE, they subjected MERS-CoV pseudoviruses to trypsin cleavage with or without pre-incubation with DPP4. The results showed that only the S1/S2 site, was accessible to proteases in the free form. In the presence of DPP4, spike molecules got cleaved at the S2’ site by trypsin. Interestingly, they found that like DPP4, Mersmab1 binding also induces conformational changes in the MERS-CoV spike to expose S2’ site to be cleaved. Combining previous studies with their recent findings they support the idea where similar to DPP4, Mersmab1 stabilizes the RBD in the standing-up position and triggers a conformational change of the spike.
They further evaluated the potential impact of different proteases on MERS-CoV pseudovirus entry. They used inhibitors specific for proprotein convertases, cell surface proteases, and lysosomal proteases and examined viral entry in each case. Interestingly, it came out that DPP4-dependent and Mersmab1-dependent MERS-CoV entries can be activated by all three proteases, the author suggested DPP4-dependent and Mersmab1-dependent MERS-CoV entries share the same pathway.
Lastly, they determined the range of Mersmab1 dosages in ADE. They found Mersmab1 blocks the DPP4-dependent viral entry pathway by outcompeting DPP4 for binding to the MERS-CoV spike. Mersmab1 first increased and then decreased (at 100ng/ml) the viral entry via the Fc receptor. likely due to saturation and blocking of indirect interaction at high concentration. Viral entry into cells expressing both DPP4 and Fc receptor first dropped, then increased, and finally dropped again. Likely, because when both DPP4 and CD32A are present on the host cell surface, Mersmab1 inhibits viral entry (by blocking the DPP4-dependent entry pathway) at low concentrations, promotes viral entry (by enhancing the CD32A-dependent entry pathway) at intermediate concentrations, and inhibits viral entry (by blocking both the DPP4- and CD32A-dependent entry pathways) at high concentrations.
Findings of the Study
Their findings reveal that RBD-specific neutralizing MAbs may mediate ADE of viruses by mimicking the functions of viral receptors only at intermediate MAb dosages. Their study suggests that ADE occurs under some specific conditions in vivo, depending on the antibody dosages, the binding affinity of the MAb for DPP4, and tissue expressions of DPP4 and Fc receptor. Moreover, the mechanism that we have identified for ADE of MERS-CoV in vitro may account for the ADE observed in vivo for other coronaviruses, such as SARS-CoV and feline coronavirus. Overall, their study reveals the complex roles of antibodies in viral entry and can guide future vaccine design and antibody-based drug therapy.
Journal Reference
- Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, He L, Chen Y, Wu J, Shi Z, Zhou Y, Du L, Li F. 2020. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol 94:e02015-19. https://doi.org/10.1128/ JVI.02015-19.
