Agrobacterium tumefaciens Mediated Transformation
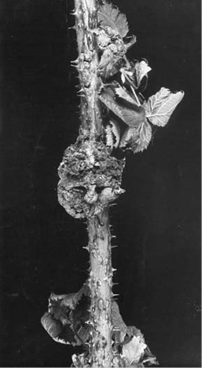
Gene transfer from bacteria to plants occurs naturally. The Ti plasmid is present in Agrobacterium tumefaciens. Agrobacterium tumefaciens infects many species of plants causing a disease known as “crown gall”.

Agrobacterium tumefaciens Mediated Transformation
Gene transfer from bacteria to plants occurs naturally. The Ti plasmid is present in Agrobacterium tumefaciens. Agrobacterium tumefaciens is a soil pathogen, a gram-negative bacterium which infects many species of plants causing a disease known as “crown gall”. It has two common species A. tumefaciens and A. rhizogenes.
Mechanism of infection at molecular level
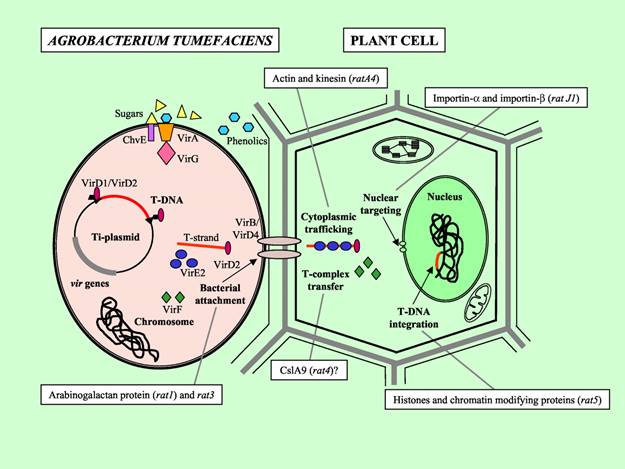
The oncogenic activity of Agrobacterium is due to the presence of a large (200-kb) tumor inducing plasmid (pTi) and tumor formation results from its infection into the host plant (dicotyledonous plants). Upon infection of the wounded plant, the bacterium transfers a small segment of DNA, the so-called T-DNA from the Ti plasmid into the plant cell.
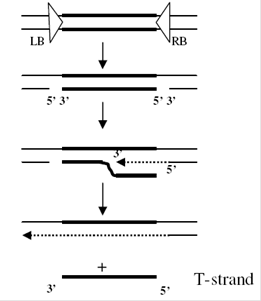
The T-DNA is then translocated into the plant nucleus and stably integrated into the chromosome. T-DNA size varies from 15 and 30 kb depending on the strain. In the Ti-plasmid itself, the T-DNA is flanked by 25-bp imperfect direct repeats known as border sequences, which are conserved between octopine and nopaline plasmids. The border sequences are not transferred intact to the plant genome, but they are involved in the transfer process. The right repeat is necessary for transfer and integration to a plant genome.
Deletion of the right-border repeat abolishes T-DNA transfer, but the left-hand border surprisingly appears to be non-essential. The right junction is rather precise, but the left junction can vary by about 100 nucleotides.

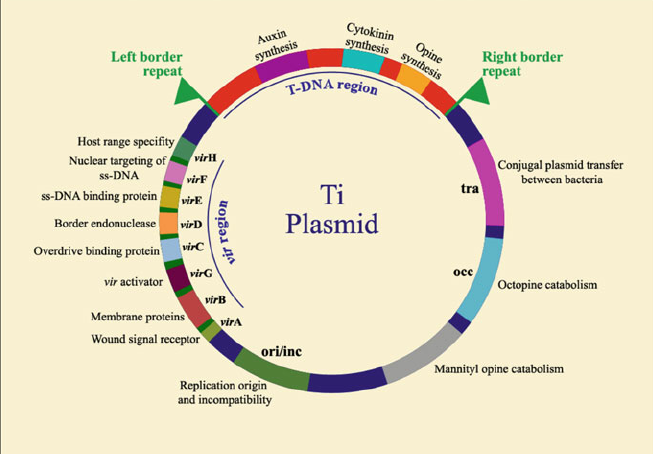
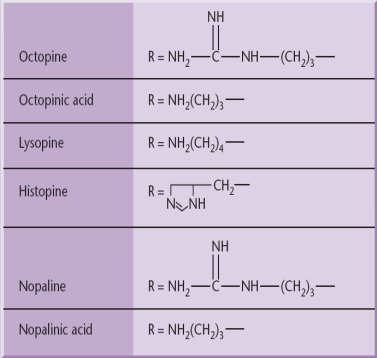
The wild type T-DNA encodes a specific set of oncogenic (onc) genes that when it is expressed in the host cell, leads to the formation of a tumor. T-DNA carries genes for phytohormone (auxin and cytokinin) and opines. The overproduction of phytohormones at the site of infection is responsible for the proliferation of wound cells into a gall (tumor) that can harbor a population of bacteria.
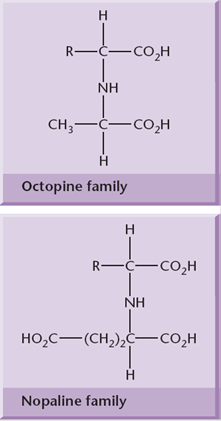
Agrobacterium strains are usually classified according to the type of opine gene present on their T-DNA: nopaline, octopine, agropine or succinamopine types. Nopaline and octopine strains are the most common. These opines are condensation products of either an amino acid and a keto acid or an amino acid and a sugar. These opines serve as a source of carbon and nitrogen for Agrobacterium.

Activation of Vir genes
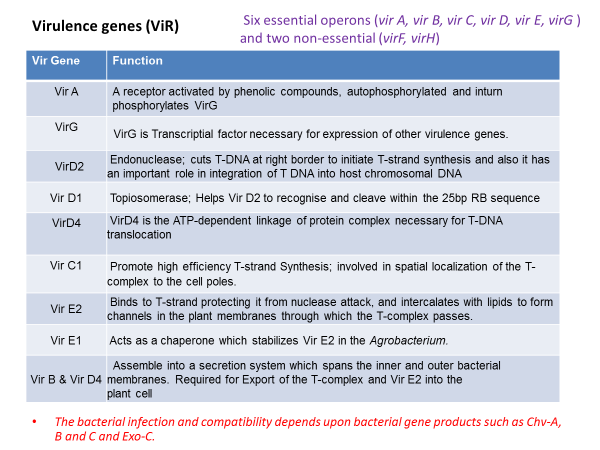
The genes responsible for T-DNA transfer are located in a separate part of the Ti-plasmid called the vir (virulence) region. This region consists of at least six major operons i.e. VirA, VirB, VirC, VirD, VirE, and VirG, which encode the virulence proteins of DNA transport.
The number of genes per operon differs: virA, virG and virF have only one gene; virE and virC have two genes while virD and virB have four and eleven genes respectively. The vir genes caused T-DNA transfer to plant cell is analogous to transfer of plasmids during bacterial conjugation.
The third element necessary for transformation are the chromosomal virulence (chv) genes which are located in the bacterial chromosome and are involved in recognition and early attachment of Agrobacterium to the plant cell.
It is those three elements that work in a synergistic fashion and enable the gene transfer by Agrobacterium. The Agrobacterium chromosomal genes i.e. chvA, chvB, pscA (exoC) and att play a key role in the recognition process of specific plant cell surface receptors.
The activation of vir genes is dependent on a two-component signal transduction system of Agrobacterium i.e. consisting of 1) a membrane sensor protein, VirA and 2) a cytoplasmic transducer protein, VirG.
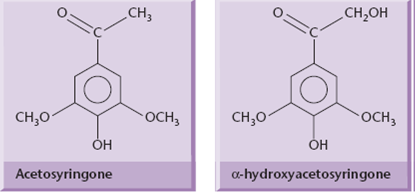
Two of these genes, virA and virG, are constitutively expressed at a low level and control the plant-induced activation of the other vir genes. VirA is a kinase that spans the inner bacterial membrane, and acts as the receptor for certain phenolic molecules (acetosyringone) that are released by wounded plant cells.
The signal transduction pathways of further vir induction begins when VirA interacts with the phenolic signal in combination with a chromosomally encoded sugar-binding protein (ChvE) and extracellular acidity resulting in VirA autophosphorylation at a specific histidine residue (His-474) of the protein.
Since the phosphorylation at this residue is unstable due to the formation of a high energy phosphate bond, VirA then transfers the phosphate group to aspartate residue 52 in VirG where it is in an active and stable form. The transphosphorylated form of VirG then binds with a conserved 12-bp sequence of a vir box enhancer element located in the promoter region of vir genes.
Generation of T-DNA complex and Transfer of T-DNA into plant cell
virB encodes proteins which produce a conjugative pilus through which the T-DNA is transferred to the plant cell. DNA transfer itself is initiated by an endonuclease formed by the products of the virD1 and virD2 genes. This introduces either single-strand nicks or a double-strand break at the 25-bp borders of the T-DNA, a process enhanced by the VirC12 and VirC2 proteins, which recognize and bind to the overdrive enhancer element present near the right border sequence, which stimulate the transfer process. The VirD2 protein remains covalently attached to the processed T-DNA via tyrosine residue 29.
Recent studies have suggested that the type of T-DNA intermediate produced (single- or double-stranded) depends on the type of Ti-plasmid, with double stranded T-DNA favored by nopaline plasmids (where the T-DNA is a single element) and single “T-strands” favored by octopine and succinopine plasmids, where the T-DNA is split into noncontiguous sections. T-strands are coated with VirE2, a single-stranded DNA binding protein.
VirE2 and virD2 contain nuclear localization signal. VirD2 protein contains a carboxyl-terminal NLS. The VirD2 protein has been proposed to protect the T-DNA against nucleases, to target the DNA to the plant cell nucleus, and to integrate it into the plant genome. After the T-strand is released, it is recruited to the Type IV Secretion System. The ssT-DNA is converted into double stranded T-DNA and integrate into the plant genome through nonhomologous recombination.
Agrobacterium based vectors
For plant transformation, the tumorous growth activity caused by Agrobacterium is prevented by deleting the oncogenes or making the genes nonfunctional by interrupting their sequence in such a way that the transformation is still effective without developing the disease. This process is called disarming. Replacing the tumor encoding region by DNA of interest does not affect the transfer of T-DNA into the host plant. This opened the way for Agrobacterium as the tool to produce transgenic plants that can express specific genes of interest.
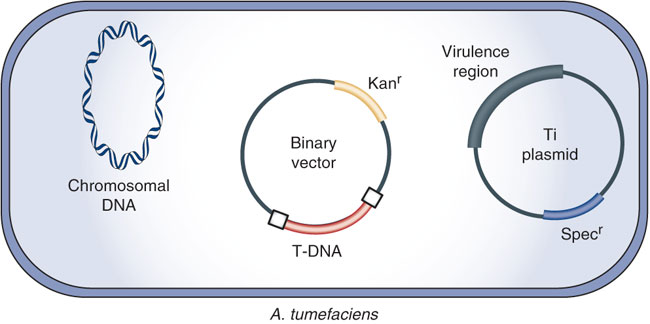
Because of Ti plasmid’s large size (about 200 kbp), direct genetic manipulation in vitro is difficult. To overcome this problem, scientists developed alternative strategies by designing several types of vectors. The most successful is the development of the binary vector system which has been commonly used for plant genetic engineering.
This strategy was based on the fact that two main components for successful Agrobacterium gene transfer, the T-DNA region and the vir genes, can reside on separate plasmids. In this system, the vir gene functions are provided by a ‘disarmed’ Ti plasmid in which the T-DNA with all the onc genes and its left (LB) and right (RB) border has been deleted, leaving the vir region and other plasmid parts intact.
A second plasmid, which can replicate and be easily manipulated in both Escherichia coli and Agrobacterium, is smaller and carries a modified T-DNA between LB and RB flanking regions containing the genes to be transferred. Typically, this plasmid has a broad host range origin of replication (ori), a bacterial gene for antibiotic resistance which serves for the selection of transformed plant cells, a marker for selection and maintenance in both E. coli and Agrobacterium. The plasmid harboring the vir gene function is termed vir helper while the plasmid harboring the T region is termed binary vector.
The Ri plasmid of A. rhizogenes and Ti plasmids are very similar, the main difference being that the transfer of the T-DNA from the Ri plasmid to plant result not in a crown gall but in hairy root disease at the site of infection.